Multilevel Models (a.k.a. Hierarchical Models)
Bayesian hierarchical models (also called multilevel models) are a statistical model written at multiple levels (hierarchical form) that estimates the parameters of the posterior distribution using the Bayesian approach. The sub-models combine to form the hierarchical model, and Bayes' theorem is used to integrate them with the observed data and to account for all the uncertainty that is present. The result of this integration is the posterior distribution, also known as an updated probability estimate, as additional evidence of the likelihood function is integrated together with the prior distribution of the parameters.
Hierarchical modeling is used when information is available at several different levels of observation units. The hierarchical form of analysis and organization helps to understand multiparameter problems and also plays an important role in the development of computational strategies.
Hierarchical models are mathematical statements that involve several parameters, so that the estimates of some parameters depend significantly on the values of other parameters. The figure below shows a hierarchical model in which there is a hyperparameter that parameterizes the parameters that are finally used to infer the posterior density of some variable of interest .
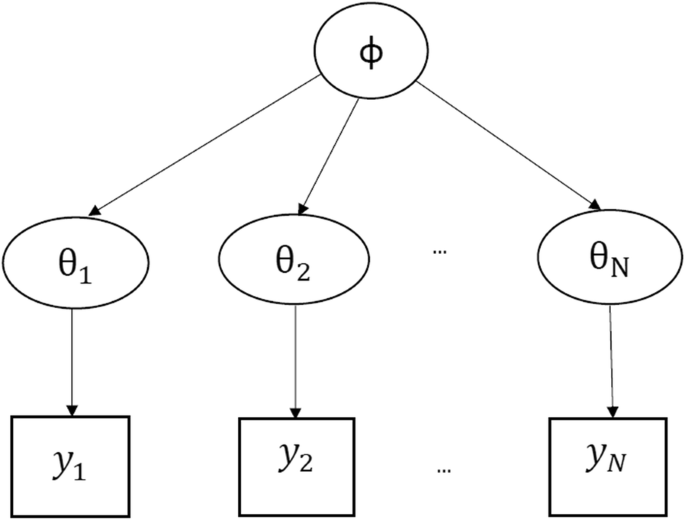
When to use Multilevel Models?
Multilevel models are particularly suitable for research projects where participant data is organized at more than one level, i.e. nested data. Units of analysis are usually individuals (at a lower level) that are nested in contextual/aggregate units (at a higher level). An example is when we are measuring the performance of individuals and we have additional information about belonging to different groups such as sex, age group, hierarchical level, educational level or housing status.
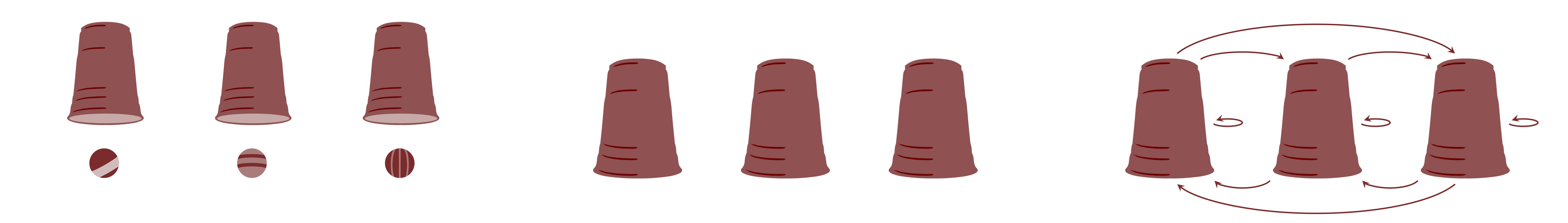
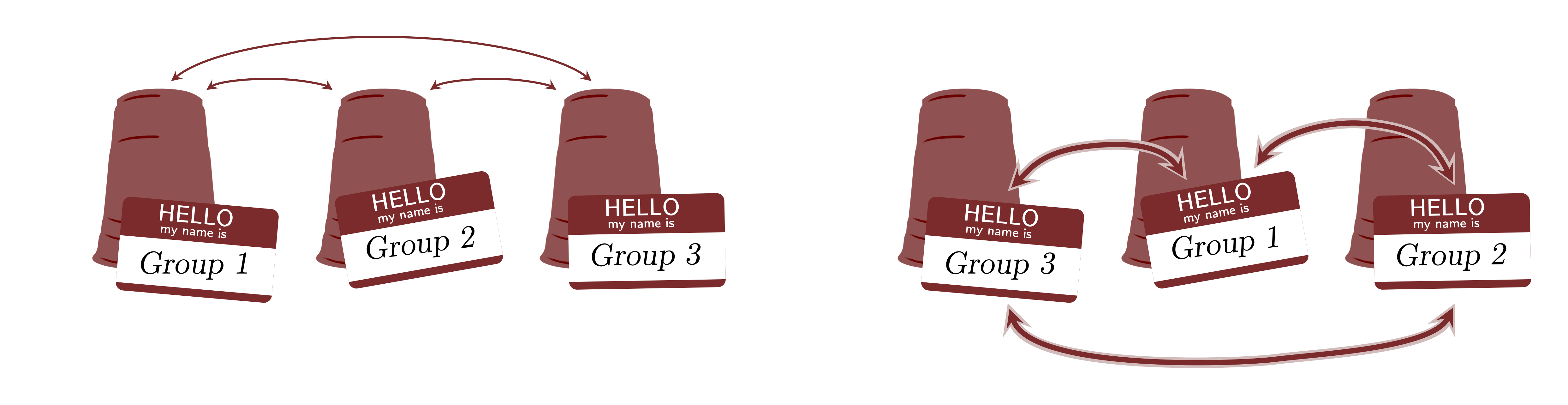
There is a main assumption that cannot be violated in multilevel models which is exchangeability (de Finetti, 1974; Nau, 2001). Yes, this is the same assumption that we discussed in 2. What is Bayesian Statistics?. This assumption assumes that groups are exchangeable. The figure below shows a graphical representation of the exchangeability. The groups shown as "cups" that contain observations shown as "balls". If in the model's inferences, this assumption is violated, then multilevel models are not appropriate. This means that, since there is no theoretical justification to support exchangeability, the inferences of the multilevel model are not robust and the model can suffer from several pathologies and should not be used for any scientific or applied analysis.
Hyperpriors
As the priors of the parameters are sampled from another prior of the hyperparameter (upper-level's parameter), which are called hyperpriors. This makes one group's estimates help the model to better estimate the other groups by providing more robust and stable estimates.
We call the global parameters as population effects (or population-level effects, also sometimes called fixed effects) and the parameters of each group as group effects (or group-level effects, also sometimes called random effects). That is why multilevel models are also known as mixed models in which we have both fixed effects and random effects.
Three Approaches to Multilevel Models
Multilevel models generally fall into three approaches:
Random-intercept model: each group receives a different intercept in addition to the global intercept.
Random-slope model: each group receives different coefficients for each (or a subset of) independent variable(s) in addition to a global intercept.
Random-intercept-slope model: each group receives both a different intercept and different coefficients for each independent variable in addition to a global intercept.
Correlated-Random-intercept-slope model: each group receives both a different intercept and different coefficients for each independent variable in addition to a global intercept; while also taking into account the correlation/covariance amongst intercept/coefficients.
Random-Intercept Model
The first approach is the random-intercept model in which we specify a different intercept for each group, in addition to the global intercept. These group-level intercepts are sampled from a hyperprior.
To illustrate a multilevel model, I will use the linear regression example with a Gaussian/normal likelihood function. Mathematically a Bayesian multilevel random-slope linear regression model is:
The priors on the global intercept , global coefficients and error , along with the Gaussian/normal likelihood on are the same as in the linear regression model. But now we have new parameters. The first are the group intercepts prior that denotes that every group has its own intercept sampled from a normal distribution centered on 0 with a standard deviation . This group intercept is added to the linear predictor inside the Gaussian/normal likelihood function. The group intercepts' standard deviation have a hyperprior (being a prior of a prior) which is sampled from a positive-constrained Cauchy distribution (a special case of the Student- distribution with degrees of freedom ) with mean 0 and standard deviation . This makes the group-level intercept's dispersions being sampled from the same parameter which allows the model to use information from one group intercept to infer robust information regarding another group's intercept dispersion and so on.
This is easily accomplished with Turing:
using Turing
using LinearAlgebra
using Statistics: mean, std
using Random: seed!
seed!(123)
@model function varying_intercept(
X, idx, y; n_gr=length(unique(idx)), predictors=size(X, 2)
)
#priors
α ~ Normal(mean(y), 2.5 * std(y)) # population-level intercept
β ~ filldist(Normal(0, 2), predictors) # population-level coefficients
σ ~ Exponential(std(y)) # residual SD
#prior for variance of random intercepts
#usually requires thoughtful specification
τ ~ truncated(Cauchy(0, 2); lower=0) # group-level SDs intercepts
αⱼ ~ filldist(Normal(0, τ), n_gr) # group-level intercepts
#likelihood
ŷ = α .+ X * β .+ αⱼ[idx]
return y ~ MvNormal(ŷ, σ^2 * I)
end;Random-Slope Model
The second approach is the random-slope model in which we specify a different slope for each group, in addition to the global intercept. These group-level slopes are sampled from a hyperprior.
To illustrate a multilevel model, I will use the linear regression example with a Gaussian/normal likelihood function. Mathematically a Bayesian multilevel random-slope linear regression model is:
Here we have a similar situation from before with the same hyperprior, but now it is a hyperprior for the the group coefficients' standard deviation prior: . This makes the group-level coefficients's dispersions being sampled from the same parameter which allows the model to use information from one group coefficients to infer robust information regarding another group's coefficients dispersion and so on.
In Turing we can accomplish this as:
@model function varying_slope(X, idx, y; n_gr=length(unique(idx)), predictors=size(X, 2))
#priors
α ~ Normal(mean(y), 2.5 * std(y)) # population-level intercept
σ ~ Exponential(std(y)) # residual SD
#prior for variance of random slopes
#usually requires thoughtful specification
τ ~ filldist(truncated(Cauchy(0, 2); lower=0), n_gr) # group-level slopes SDs
βⱼ ~ filldist(Normal(0, 1), predictors, n_gr) # group-level standard normal slopes
#likelihood
ŷ = α .+ X * βⱼ * τ
return y ~ MvNormal(ŷ, σ^2 * I)
end;Random-Intercept-Slope Model
The third approach is the random-intercept-slope model in which we specify a different intercept and slope for each group, in addition to the global intercept. These group-level intercepts and slopes are sampled from hyperpriors.
To illustrate a multilevel model, I will use the linear regression example with a Gaussian/normal likelihood function. Mathematically a Bayesian multilevel random-intercept-slope linear regression model is:
Here we have a similar situation from before with the same hyperpriors, but now we fused both random-intercept and random-slope together.
In Turing we can accomplish this as:
@model function varying_intercept_slope(
X, idx, y; n_gr=length(unique(idx)), predictors=size(X, 2)
)
#priors
α ~ Normal(mean(y), 2.5 * std(y)) # population-level intercept
σ ~ Exponential(std(y)) # residual SD
#prior for variance of random intercepts and slopes
#usually requires thoughtful specification
τₐ ~ truncated(Cauchy(0, 2); lower=0) # group-level SDs intercepts
τᵦ ~ filldist(truncated(Cauchy(0, 2); lower=0), n_gr) # group-level slopes SDs
αⱼ ~ filldist(Normal(0, τₐ), n_gr) # group-level intercepts
βⱼ ~ filldist(Normal(0, 1), predictors, n_gr) # group-level standard normal slopes
#likelihood
ŷ = α .+ αⱼ[idx] .+ X * βⱼ * τᵦ
return y ~ MvNormal(ŷ, σ^2 * I)
end;In all of the models so far, we are using the MvNormal construction where we specify both a vector of means (first positional argument) and a covariance matrix (second positional argument). Regarding the covariance matrix σ^2 * I, it uses the model's errors σ, here parameterized as a standard deviation, squares it to produce a variance parameterization, and multiplies by I, which is Julia's LinearAlgebra standard module implementation to represent an identity matrix of any size.
Correlated-Random-Intercept-Slope Model
The third approach is the correlated-random-intercept-slope model in which we specify a different intercept and slope for each group, in addition to the global intercept. These group-level intercepts and slopes are sampled from hyperpriors. Finally, we also model the correlation/covariance amongst the intercepts and slopes. We assume that they are not independent anymore, rather they are correlated.
In order to model the correlation between parameters, we need a prior on correlation/covariance matrices. There are several ways to define priors on covariance matrices. In some ancient time, "Bayesian elders" used distributions such as the Wishart distribution and the Inverse Wishart distribution. However, priors on covariance matrices are not that intuitive, and can be hard to translate into real-world scenarios. That's why I much prefer to construct my covariances matrices from a correlation matrix and a vector of standard deviations. Remember that every covariance matrix can be decomposed into:
where is a correlation matrix with s in the diagonal and the off-diagonal elements between -1 e 1 . is a vector composed of the variables' standard deviation from (is is the 's diagonal).
These are much more intuitive and easy to work with, specially if you are not working together with some "Bayesian elders". This approach leaves us with the need to specify a prior on correlation matrices. Luckily, we have the LKJ distribution: distribution for positive definite matrices with unit diagonals (exactly what a correlation matrix is).
To illustrate a multilevel model, I will use the linear regression example with a Gaussian/normal likelihood function. Mathematically a Bayesian multilevel correlated-random-intercept-slope linear regression model is:
We don't have any s. If we want a varying intercept, we just insert a column filled with s in the data matrix . Mathematically, this makes the column behave like an "identity" variable (because the number in the multiplication operation is the identity element. It maps keeping the value of intact) and, consequently, we can interpret the column's coefficient as the model's intercept.
In Turing we can accomplish this as:
using PDMats
@model function correlated_varying_intercept_slope(
X, idx, y; n_gr=length(unique(idx)), predictors=size(X, 2)
)
#priors
Ω ~ LKJCholesky(predictors, 2.0) # Cholesky decomposition correlation matrix
σ ~ Exponential(std(y))
#prior for variance of random correlated intercepts and slopes
#usually requires thoughtful specification
τ ~ filldist(truncated(Cauchy(0, 2); lower=0), predictors) # group-level SDs
γ ~ filldist(Normal(0, 5), predictors, n_gr) # matrix of group coefficients
#reconstruct Σ from Ω and τ
Σ_L = Diagonal(τ) * Ω.L
Σ = PDMat(Cholesky(Σ_L + 1e-6 * I)) # numerical instability
#reconstruct β from Σ and γ
β = Σ * γ
#likelihood
return y ~ arraydist([Normal(X[i, :] ⋅ β[:, idx[i]], σ) for i in 1:length(y)])
end;In the correlated_varying_intercept_slope model, we are using the efficient Cholesky decomposition version of the LKJ prior with LKJCholesky. We put priors on all group coefficients γ and reconstruct both the covariance matrix Σ and the coefficient matrix β.
Example - Cheese Ratings
For our example, I will use a famous dataset called cheese (Boatwright, McCulloch & Rossi, 1999), which is data from cheese ratings. A group of 10 rural and 10 urban raters rated 4 types of different cheeses (A, B, C and D) in two samples. So we have observations and 4 variables:
cheese: type of cheese fromAtoDrater: id of the rater from1to10background: type of rater, eitherruralorurbany: rating of the cheese
OK, let's read our data with CSV.jl and output into a DataFrame from DataFrames.jl:
using DataFrames
using CSV
using HTTP
url = "https://raw.githubusercontent.com/storopoli/Bayesian-Julia/master/datasets/cheese.csv"
cheese = CSV.read(HTTP.get(url).body, DataFrame)
describe(cheese)4×7 DataFrame
Row │ variable mean min median max nmissing eltype
│ Symbol Union… Any Union… Any Int64 DataType
─────┼───────────────────────────────────────────────────────────────
1 │ cheese A D 0 String1
2 │ rater 5.5 1 5.5 10 0 Int64
3 │ background rural urban 0 String7
4 │ y 70.8438 33 71.5 91 0 Int64As you can see from the describe() output, the mean cheese ratings is around 70 ranging from 33 to 91.
In order to prepare the data for Turing, I will convert the Strings in variables cheese and background to Ints. Regarding cheese, I will create 4 dummy variables one for each cheese type; and background will be converted to integer data taking two values: one for each background type. My intent is to model background as a group both for intercept and coefficients. Take a look at how the data will look like for the first 5 observations:
for c in unique(cheese[:, :cheese])
cheese[:, "cheese_$c"] = ifelse.(cheese[:, :cheese] .== c, 1, 0)
end
cheese[:, :background_int] = map(cheese[:, :background]) do b
if b == "rural"
1
elseif b == "urban"
2
else
missing
end
end
first(cheese, 5)5×9 DataFrame
Row │ cheese rater background y cheese_A cheese_B cheese_C cheese_D background_int
│ String1 Int64 String7 Int64 Int64 Int64 Int64 Int64 Int64
─────┼───────────────────────────────────────────────────────────────────────────────────────────
1 │ A 1 rural 67 1 0 0 0 1
2 │ A 1 rural 66 1 0 0 0 1
3 │ B 1 rural 51 0 1 0 0 1
4 │ B 1 rural 53 0 1 0 0 1
5 │ C 1 rural 75 0 0 1 0 1Now let's us instantiate our model with the data. Here, I will specify a vector of Ints named idx to represent the different observations' group memberships. This will be used by Turing when we index a parameter with the idx, e.g. αⱼ[idx].
X = Matrix(select(cheese, Between(:cheese_A, :cheese_D)));
y = cheese[:, :y];
idx = cheese[:, :background_int];The first model is the varying_intercept:
model_intercept = varying_intercept(X, idx, y)
chain_intercept = sample(model_intercept, NUTS(), MCMCThreads(), 1_000, 4)
println(DataFrame(summarystats(chain_intercept)))9×8 DataFrame
Row │ parameters mean std mcse ess_bulk ess_tail rhat ess_per_sec
│ Symbol Float64 Float64 Float64 Float64 Float64 Float64 Float64
─────┼───────────────────────────────────────────────────────────────────────────────────────
1 │ α 70.7489 5.02573 0.176587 1049.21 944.855 1.00255 19.8948
2 │ β[1] 2.91501 1.30963 0.0257346 2596.92 2763.9 1.00097 49.2419
3 │ β[2] -10.632 1.34056 0.028425 2224.99 2724.4 1.00189 42.1895
4 │ β[3] 6.52144 1.3159 0.0265587 2450.36 2667.92 1.00052 46.463
5 │ β[4] 1.09178 1.31719 0.0264376 2475.97 2627.54 1.00117 46.9485
6 │ σ 7.3913 0.449209 0.00807096 3127.77 2530.83 1.00102 59.3078
7 │ τ 6.11784 5.54993 0.152948 1863.81 1849.7 1.00172 35.341
8 │ αⱼ[1] -3.39662 4.92084 0.174051 1057.29 929.725 1.00312 20.0479
9 │ αⱼ[2] 3.63474 4.95267 0.175295 1054.17 920.198 1.00331 19.9889
Here we can see that the model has a population-level intercept α along with population-level coefficients βs for each cheese dummy variable. But notice that we have also group-level intercepts for each of the groups αⱼs. Specifically, αⱼ[1] are the rural raters and αⱼ[2] are the urban raters.
Now let's go to the second model, varying_slope:
model_slope = varying_slope(X, idx, y)
chain_slope = sample(model_slope, NUTS(), MCMCThreads(), 1_000, 4)
println(DataFrame(summarystats(chain_slope)))12×8 DataFrame
Row │ parameters mean std mcse ess_bulk ess_tail rhat ess_per_sec
│ Symbol Float64 Float64 Float64 Float64 Float64 Float64 Float64
─────┼────────────────────────────────────────────────────────────────────────────────────────
1 │ α 71.6225 6.12929 0.80038 70.2758 24.8834 1.0542 1.3079
2 │ σ 7.98871 0.435528 0.0142789 879.095 1907.61 1.00481 16.3607
3 │ τ[1] 5.95148 5.05219 0.283585 297.117 1630.98 1.01456 5.52961
4 │ τ[2] 7.13182 7.54656 1.22127 120.798 26.97 1.03962 2.24817
5 │ βⱼ[1,1] 0.199936 0.839602 0.0512732 282.448 223.989 1.01652 5.25661
6 │ βⱼ[2,1] -0.933251 1.06721 0.0676919 231.737 252.835 1.01661 4.31283
7 │ βⱼ[3,1] 0.522532 0.883634 0.035598 636.854 1246.34 1.02562 11.8524
8 │ βⱼ[4,1] 0.0664401 0.797414 0.0514516 278.881 154.864 1.02163 5.19022
9 │ βⱼ[1,2] 0.290895 0.843615 0.0732187 135.896 84.0176 1.03096 2.52915
10 │ βⱼ[2,2] -0.853134 1.05917 0.0568238 415.139 467.237 1.01995 7.7261
11 │ βⱼ[3,2] 0.563184 0.876935 0.0375404 474.159 818.21 1.01366 8.82451
12 │ βⱼ[4,2] 0.124051 0.787212 0.0525178 224.833 143.946 1.018 4.18434
Here we can see that the model has still a population-level intercept α. But now our population-level coefficients βs are replaced by group-level coefficients βⱼs along with their standard deviation τᵦs. Specifically βⱼ's first index denotes the 4 dummy cheese variables' and the second index are the group membership. So, for example βⱼ[1,1] is the coefficient for cheese_A and rural raters (group 1).
Now let's go to the third model, varying_intercept_slope:
model_intercept_slope = varying_intercept_slope(X, idx, y)
chain_intercept_slope = sample(model_intercept_slope, NUTS(), MCMCThreads(), 1_000, 4)
println(DataFrame(summarystats(chain_intercept_slope)))15×8 DataFrame
Row │ parameters mean std mcse ess_bulk ess_tail rhat ess_per_sec
│ Symbol Float64 Float64 Float64 Float64 Float64 Float64 Float64
─────┼───────────────────────────────────────────────────────────────────────────────────────
1 │ α 70.8973 7.06973 0.226273 1080.56 988.602 1.00354 13.1307
2 │ σ 7.07716 0.403315 0.00763609 2791.06 2389.42 1.00236 33.9161
3 │ τₐ 6.19068 5.86481 0.187349 1286.35 1007.7 1.00342 15.6313
4 │ τᵦ[1] 6.19739 5.39243 0.202217 646.032 1159.5 1.00601 7.85039
5 │ τᵦ[2] 6.24244 5.30258 0.215136 598.407 1527.72 1.00463 7.27167
6 │ αⱼ[1] -3.62952 5.05254 0.176822 1253.95 761.597 1.00336 15.2377
7 │ αⱼ[2] 3.42934 5.05582 0.176181 1271.82 839.894 1.00348 15.4548
8 │ βⱼ[1,1] 0.273658 0.779529 0.0188846 1727.94 1703.37 1.00193 20.9975
9 │ βⱼ[2,1] -0.88519 1.07495 0.0402493 794.926 375.533 1.00615 9.6597
10 │ βⱼ[3,1] 0.540418 0.897065 0.0325307 849.719 418.537 1.00343 10.3255
11 │ βⱼ[4,1] 0.119311 0.761924 0.0183527 1730.92 1000.11 1.00092 21.0336
12 │ βⱼ[1,2] 0.230796 0.819673 0.0210441 1571.83 1681.27 1.00307 19.1004
13 │ βⱼ[2,2] -0.916136 1.02804 0.0319528 1027.79 2087.51 1.00208 12.4894
14 │ βⱼ[3,2] 0.577911 0.873303 0.0269948 1094.9 1738.12 1.00304 13.3049
15 │ βⱼ[4,2] 0.115502 0.784112 0.0186258 1758.58 1981.55 1.0031 21.3698
Now we have fused the previous model in one. We still have a population-level intercept α. But now we have in the same model group-level intercepts for each of the groups αⱼs and group-level along with their standard deviation τₐ. We also have the coefficients βⱼs with their standard deviation τᵦs. The parameters are interpreted exactly as the previous cases.
Now let's go to the fourth model, varying_intercept_slope. Here we are going to add a columns of into the data matrix X:
X_correlated = hcat(fill(1, size(X, 1)), X)
model_correlated = correlated_varying_intercept_slope(X_correlated, idx, y)
chain_correlated = sample(model_correlated, NUTS(), MCMCThreads(), 1_000, 4)
println(DataFrame(summarystats(chain_correlated)))31×8 DataFrame
Row │ parameters mean std mcse ess_bulk ess_tail rhat ess_per_sec
│ Symbol Float64 Float64 Float64 Float64 Float64 Float64 Float64
─────┼───────────────────────────────────────────────────────────────────────────────────────────────
1 │ Ω.L[1,1] 1.0 0.0 NaN NaN NaN NaN NaN
2 │ Ω.L[2,1] 0.0499316 0.31051 0.00698543 1975.54 2314.64 1.00168 1.52283
3 │ Ω.L[3,1] -0.454537 0.249772 0.00571052 1977.49 2262.51 1.00096 1.52433
4 │ Ω.L[4,1] 0.308835 0.25287 0.00599969 1843.28 2199.73 1.00014 1.42088
5 │ Ω.L[5,1] -0.125632 0.307712 0.00654629 2239.37 2376.57 1.00314 1.72619
6 │ Ω.L[2,2] 0.946451 0.0731193 0.00168809 2176.21 2182.87 1.00018 1.67751
7 │ Ω.L[3,2] 0.00316296 0.326756 0.00512707 4010.69 2567.33 1.0003 3.0916
8 │ Ω.L[4,2] 0.0351896 0.354115 0.00527131 4546.36 3111.1 1.00178 3.50452
9 │ Ω.L[5,2] 0.0261588 0.366052 0.00538327 4345.52 2064.09 1.00291 3.3497
10 │ Ω.L[3,3] 0.775999 0.148672 0.00348151 1845.72 2482.95 1.00082 1.42276
11 │ Ω.L[4,3] -0.0330662 0.349329 0.00604209 3242.42 2090.97 1.00099 2.49939
12 │ Ω.L[5,3] 0.0374594 0.360744 0.00516219 4901.98 2431.25 1.00176 3.77865
13 │ Ω.L[4,4] 0.753896 0.150472 0.00357772 1850.93 2236.89 1.00165 1.42677
14 │ Ω.L[5,4] 0.0116934 0.349883 0.00506856 4664.43 2772.27 1.00128 3.59553
15 │ Ω.L[5,5] 0.684545 0.179677 0.00469272 1541.5 1667.6 1.00377 1.18825
16 │ σ 7.08991 0.392342 0.00609954 4281.99 2994.66 1.00037 3.30073
17 │ τ[1] 3.82687 0.792461 0.0243249 1071.64 1274.54 1.00157 0.82606
18 │ τ[2] 0.515217 0.457244 0.0118942 1162.98 1701.28 1.00367 0.896471
19 │ τ[3] 1.57042 0.675595 0.0194601 1075.05 1066.81 1.00327 0.82869
20 │ τ[4] 0.79098 0.502001 0.0137349 1042.75 683.387 1.0037 0.803794
21 │ τ[5] 0.536124 0.435909 0.0118638 1106.15 1058.94 1.00122 0.852668
22 │ γ[1,1] 5.1869 2.19555 0.0618344 1249.75 1539.01 1.00063 0.963357
23 │ γ[2,1] -0.104914 4.6473 0.0843589 3038.11 2765.66 0.999955 2.3419
24 │ γ[3,1] -2.26726 3.24333 0.0677281 2307.05 2102.85 1.00399 1.77836
25 │ γ[4,1] 0.786036 4.18682 0.0682086 3757.86 2648.24 1.00005 2.89671
26 │ γ[5,1] 0.317867 4.53047 0.067744 4456.83 2826.46 1.0041 3.43551
27 │ γ[1,2] 5.68517 2.38212 0.0679177 1209.5 1530.76 1.00015 0.932333
28 │ γ[2,2] 0.406946 4.69823 0.0761723 3785.93 2816.48 1.00131 2.91835
29 │ γ[3,2] -2.43749 3.41662 0.0695933 2451.7 2481.02 1.00083 1.88987
30 │ γ[4,2] 1.40997 4.31934 0.0742324 3389.91 2550.09 1.00076 2.61308
31 │ γ[5,2] -0.565495 4.69425 0.0822518 3281.19 2340.3 1.00369 2.52928
We can see that we also get all of the Cholesky factors of the correlation matrix Ω which we can transform back into a correlation matrix by doing Ω = Ω.L * Ω.L'.
References
Boatwright, P., McCulloch, R., & Rossi, P. (1999). Account-level modeling for trade promotion: An application of a constrained parameter hierarchical model. Journal of the American Statistical Association, 94(448), 1063–1073.
de Finetti, B. (1974). Theory of Probability (Volume 1). New York: John Wiley & Sons.
Nau, R. F. (2001). De Finetti was Right: Probability Does Not Exist. Theory and Decision, 51(2), 89–124. https://doi.org/10.1023/A:1015525808214